User Manual
Overview
G-quadruplexes (G4s) play crucial roles in key biological functions, including transcription, replication, telomere maintenance, and genomic instability, and have been an important component of organismal evolution.
Quadrupia is a comprehensive database of G4 sequences for a large collection of reference genomes from GenBank and RefSeq, enabling their study across the tree of life. Each genome has been annotated using two distinct methods (regular expressions and G4hunter) for the detection of G4 sequences. In addition, the database contains a curated list of G4 sequence clusters, created using the linclust algorithm of MMseqs2.
Through Quadrupia, users can:
- Browse the G4 sequences of more than 100,000 reference genomes from all organism groups (Bacteria, Archaea, Eukaryota and Viruses).
- Explore a curated list of 319,784 sequence clusters.
- Perform sequence-based, HMM-based and motif-based queries against the database's contents.
Availability
Quadrupia is available through: https://pavlopoulos-lab.org/quadrupia.
How to use this website
You can explore our database using the navigation guide at the top of the page:
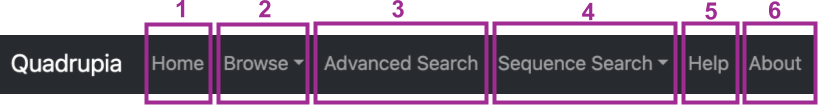
-
By clicking on "Home" (1), you'll be directed to a page containing a brief description of G quadruplexes as a biological concept along with key information about Quadrupia. Additionally, you can view the overall statistics of the database contents and learn how to properly cite it.
-
By clicking on "Browse" (2), you can choose between "Genomes" or "G4 Clusters". You will be directed to a dedicated page where you can explore the G4 sequences of over 100,000 reference genomes from all organism groups (Bacteria, Archaea, Eukaryota, and Viruses). You can browse by individual genome or by clusters, where genomes containing G4 sequences are organized.
-
Clicking on "Advanced Search" (3), you can explore a curated list of 319,784 sequence clusters, either at the genome or cluster level.
-
You can perform sequence-based, HMM-based, and motif-based queries against the database's contents by clicking on "Sequence Search" (4).
-
The "Help" page (5) will guide you on how to use and explore all the utilities of the database, whereas the "About" page (6) will provide more information about the team and how to cite the database.
How to use this help manual
Topics in the Manual are divided in separate tabs, accessible through header buttons at the top of the page. Click on any of these header buttons to navigate to its respective section.
Long sections are divided into subsections, that can be scrolled up and down. At the end of each subsection a link exists, labeled [Back to top]. Clicking on it will return you to the top of the section
Browsing Data
Browsing data in Quadrupia is available through the Browse menu in the navigation bar. By clicking on Browse, you can select between Genomes and G4 Clusters.
Browse Genomes
When you select Genomes, you will be directed to the Browse Genomes page. Here, you can explore the G4 sequences of over 100,000 reference genomes from all organism groups (Bacteria, Archaea, Eukaryota, and Viruses) presented in an interactive table with four columns.
- You can navigate each column using the grey header at the top.
- You can filter the table content for specific entries in each category (Accession Assembly, organism, taxonomic group, or number of G4 sequences) by typing in the search bar in each column.
- You can also sort the results presented in the page by each column using the up and down arrows next to the column name.
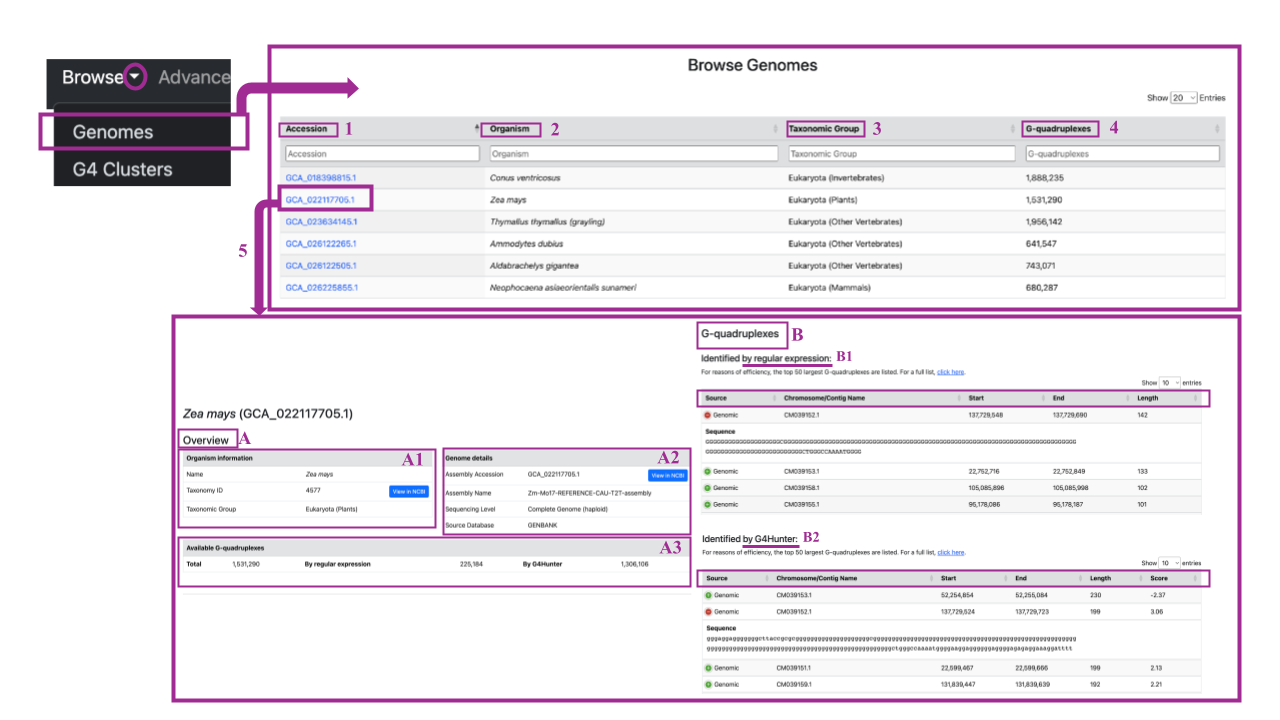
By clicking on a specific accession ID (as shown in example (5)), you will be redirected to a dedicated page for that genome accession ID. Here you can find:
(A) Overview Table:
- A1) Information about the organism (name, taxon ID with a link to NCBI, and taxonomic group).
- A2) Genome details (Assembly Accession with a link to NCBI, Assembly Name, Sequencing level -Complete Genome or Chromosome with reference to Haploid or Diploid and Source database
- A3) Available G-quadruplexes section, showing the total number of G4 sequences detected in this assembly accession, with details on how many were identified by regular expression and G4 Hunter.
By scrolling down you can find,
(B) G-quadruplexes Section:
- (B1) Top 50 G4 sequences identified by regular expression: Includes details on the Source (Genomic, CDS or RNA), Chromosome/contig name, Starting and Ending positions, and Sequence length. Each row starts with a green button that, when clicked, opens a new tab containing the sequence itself. For the full list, click "click here" at the top of the table.
- (B2) Top 50 G4 sequences identified by G4 Hunter: Similar structure to the previous table, including the Source, Chromosome/contig name, Starting and Ending positions, Sequence length, and an additional column for the G4 Hunter coinfidence score.
Browse G4 clusters
By selecting the G4 Clusters option under Browse, you can explore a curated list of G4 sequence clusters. These clusters are created using the linclust algorithm of MMseqs2, allowing users to explore similar G4 sequences grouped together.
When you click on Browse → G4 Clusters, you will be redirected to a new page containing a table with all the clusters generated from the G4 database sequences.
- You can navigate each column using the information in the grey header at the top.
- You can filter the table content for specific entries in each category by typing in the search bar in each column.
- You can also sort the table by each column using the up and down arrows.
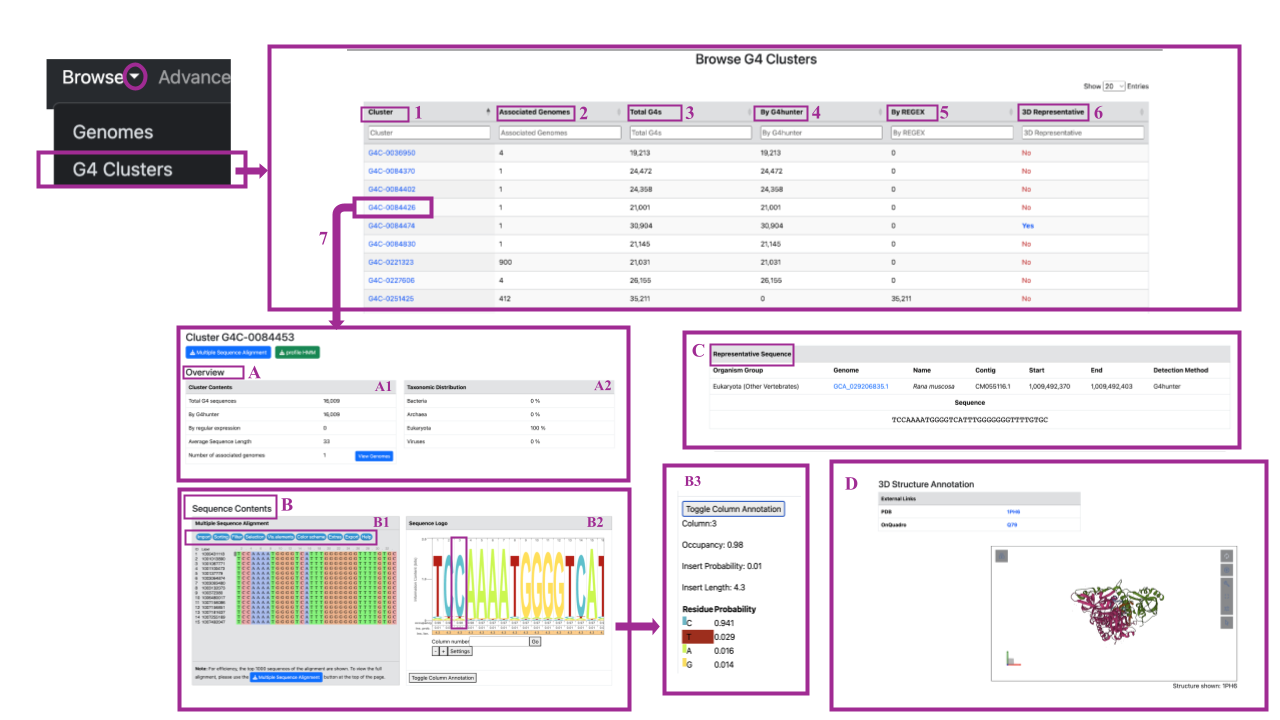
By clicking on a cluster name in Column 1 of the table, you will be redirected to a new page dedicated to that specific cluster (as shown in the example (7)). You can easily obtain the Multiple Sequence Alignment and HMM profile for each cluster by simply clicking on the respective button (colored in blue and green) located below the name of the cluster. By scrolling down, you can navigate through the page, which includes four sections (A-D):
Section A: Overview This section contains two tables (A1, A2):
Table A1: Cluster Contents This table provides the following information:
- Total G4 Sequences: The total number of G4 sequences contained in the cluster.
- By G4Hunter: The number of G4 sequences extracted by the G4 Hunter method.
- By Regular Expression: The number of G4 sequences extracted by regular expression.
- Average Sequence Length: The average length of the G4 sequences in the cluster.
- Associated Genomes: The number of genomes from which the G4 sequences in the cluster are derived. You have the option to view these genomes by clicking on the blue button on the left.
Table A2: Taxonomic Distribution This table provides information about the taxonomic annotation of G4 sequences in the cluster. You can find the percentage of G4 sequences annotated as Bacteria, Archaea, Eukaryota, and/or Viruses. It is important to note that there are clusters containing only G4 sequences identified by G4 Hunter, clusters with sequences identified by regular expression, and mixed clusters. Additionally, there are clusters that come from only one taxonomic domain and mixed clusters.
Section B: Sequence Contents This section contains two tables (B1, B2):
Table B1: Multiple Sequence Alignment In this table, you can find the multiple sequence alignment of the top 1,000 G-quadruplexes within the cluster, generated by MAFFT and displayed using Jalview. If you wish to view the full alignment, you can download it by clicking on the blue button at the bottom of the table. The alignment includes a column with sequence IDs, labels, and the aligned sequences. In the mauve box at the top of the alignment, you will find various options, features, and information accessible via blue buttons. These allow you to:
- Import/Export Files: Upload new alignments or download existing ones.
- Help Page: Access help documentation for guidance on using the tool.
- Sort: Sort the sequence IDs, Labels or Gaps by various parameters.
- Filter: Hide columns or sequences based on specific criteria.
- Selection: Identify motifs within the sequences and invert columns and/or rows.
- Vis. Elements: Show meta-information, an overview panel, sequence logo, gap weight and other.
- Color Scheme: Change the color scheme of the alignment to suit your preferences.
Table B2: Sequence logo (HMM profile)
Here, you can find an interactive sequence logo for the cluster. You can navigate through the image by scrolling left or right, and clicking on the image allows you to copy or save it. The height of each residue in the logo corresponds to its frequency, providing a visual representation of the sequence conservation within the cluster.
The interactive sequence logo includes several useful features:
- Navigation: Use the scrollbar to move through the sequence logo.
- Arithmetic Meter: Located at the top of the image, this helps you quickly find specific columns.
- Overview Panel: At the bottom of the image, you can see an overview of occupancy, insert probability, and insert length for each column.
To explore specific columns in more detail, type the column number in the search gap labeled "Column" or click on a residue. A new tab will open, providing detailed information about the selected residue/column, including occupancy, insert probability, and length, along with a table showing residue probability. To close the detailed view tab, click the "toggle column annotation" button.
Section C: Representative Sequence
In this section, you can find the representative sequence of the selected cluster, which has been extracted using the linclust algorithm. The upper part of the table provides information about the sequence, including:
- Organism Group
- Genome (as a link to each in the database)
- Name of the Organism Genome
- Contig
- Start Position of the Sequence in the Genome
- End Position of the Sequence in the Genome
- Detection Method of the G4 Representative Sequence (G4 Hunter of Regular Expression)
The lower part of the table displays the sequence itself.
Section D: 3D structure Annotation (if it is available):
You can access the 3D structure of the representative sequence of the cluster here. These structures are sourced from other repositories of structural G quadruplexes (PDB, OnQuadro) and you can navigate to them using the provided external links. The image of the 3D structure is interactive, allowing you to zoom in and out, rotate the structure, and select specific residues. Additionally, it offers an animation feature that enables you to control parameters such as duration or mode. You can even take screenshots of the structure.
Advanced Search
Youc can perform complex data queries by selecting Advanced Search from the navigation bar. An input form with two options will be presented. You can select between Genome Assemblies (A) and G-quadruplex Clusters (B).
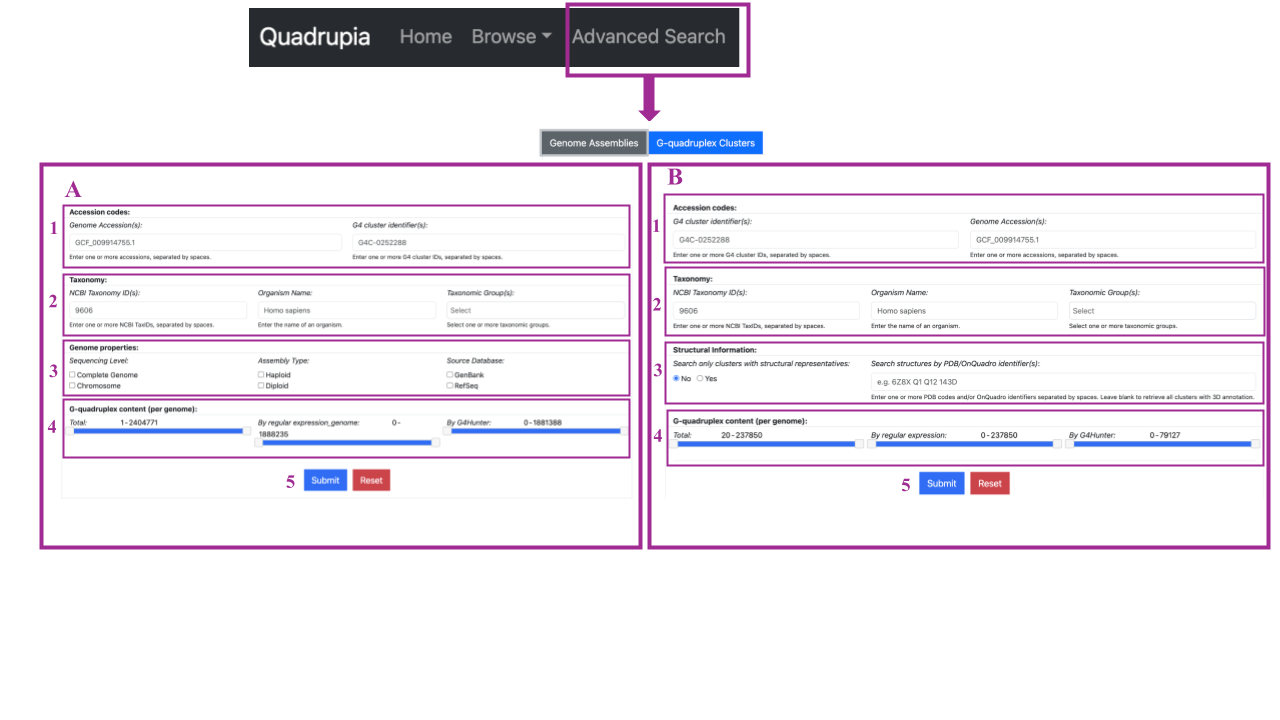
Genome Assemblies Search (A)
Here, you can search against genome assemblies using four (4) search chunks..
- Accession Codes: You can search by typing one or more genome accessions separated by space or one or more G4 cluster identifiers.
- Taxonomy: Search by typing NCBI Taxonomy IDs (one or more separated by comma), organism name (only one), and taxonomic groups of genome accessions of interest (one or more separated by comma).
- Genome Properties: You can search for specific genome accessions by providing information about genome properties such as sequence level (complete genome or chromosome), assembly type (haploid or diploid), and source database (GenBank or RefSeq).
- G-quadruplexes Content (Per Genome): Define the preferred range of G4 sequences detected per genome, and also specify the range of them that are coming from specific detection methods (G4 Hunter or Regular Expression).
By clicking Submit (5) you submit your search parameters and a new tab with your search results will be presented. Clicking on “Reset” all the filters are removed and a new search can be performed.
G-quadruplex Clusters Search (B)
Here, you can search against genome assemblies using four (4) search chunks..
- Accession Codes: You can search by typing one or more G4 cluster identifiers separated by space or one or more genome accessions.
- Taxonomy: Search by typing NCBI Taxonomy IDs (one or more separated by comma), organism name (only one), and taxonomic groups of G4 cluster of interest (one or more separated by comma).
- Structural Information: You can select only clusters with structural representation or, more specific, specific structures by PDB/ OnQuadro by typing the identifier(s) of interest. You can enter one ore more identifier separated by spaces.
- G-quadruplexes Content (Per Cluster): Define the preferred range of G4 sequences detected per cluster, and also specify the range of them that are coming from specific detection methods (G4 Hunter or Regular Expression). Note that we only consider clusters with 20 or more members.
Sequence Search
Quadrupia offers a number of options for performing sequence-based queries on its contents, accessible throug the Sequence Search menu in the navigation bar:
- G4 Sequences: Search G4 sequences using pairwise alignments with BLAST
- G4 Clusters: Search G4 clusters with profile Hidden Markov Model (pHMM)-based queries and HMMER.
- Motif Search: Search G4 sequences using custom sequence motifs.
G4 Sequence Search (BLAST)
Selecting Browse → G4 Sequences will redirect you to the "G4 Sequence Search (BLAST)" page. Here, you can search by structure in two steps:
-
Step 1 (A): You can either input or paste one or more DNA sequences, or upload a file by clicking the "Choose File" button in the lower part of the search box.
-
Step 2 (B): You can adjust BLAST search parameters by selecting:
- The reference G-quadruplex set: Specify your search against sequences detected only by regular expression, only by G4 Hunter, or against all sequences (which may be very slow).
- Search method: Choose the BLAST search method between blastn-short (for short sequences) and blastn (for sequences longer than 30 nucleotides).
- Word size: refers to the length of the sequence segment (or "word") used as the initial search unit. BLAST identifies exact or similar word matches between the query and database sequences to find "hot-spots" for initiating alignments. In nucleotide-nucleotide searches (blastn), an exact match of the entire word is required to start an extension. The sensitivity and speed of the search can be adjusted by increasing or decreasing the word size: smaller word sizes increase sensitivity but slow down the search, while larger word sizes decrease sensitivity but speed up the search.decrease sensitivity but speed up the search.
- Match/Mismatch score: Set the match/mismatch score threshold for your BLAST search, which influences the scoring of alignments based on nucleotide matches and mismatches.
- Gap costs: Define the penalties for opening and extending gaps in the alignment, which affect the overall alignment score by discouraging or allowing gaps.
- Expect Threshold: Set the E-value threshold for reporting matches. Lower E-values indicate more stringent matches.
You can also try examples by clicking on the grey button labeled Load Example. An example sequence will automatically load into the box, allowing you to experiment with different search parameters and become familiar with the search panel.
After the sequence of interest has been loaded in the search panel and the search parameters have been defined, you can submit your selections by clicking the blue Submit button at the bottom of the page, which will redirect you to the results page. If you selected the Open results in a new tab option, your results will be displayed in a new tab. By clicking the red Reset button, you can clear all your selections and start a new search.
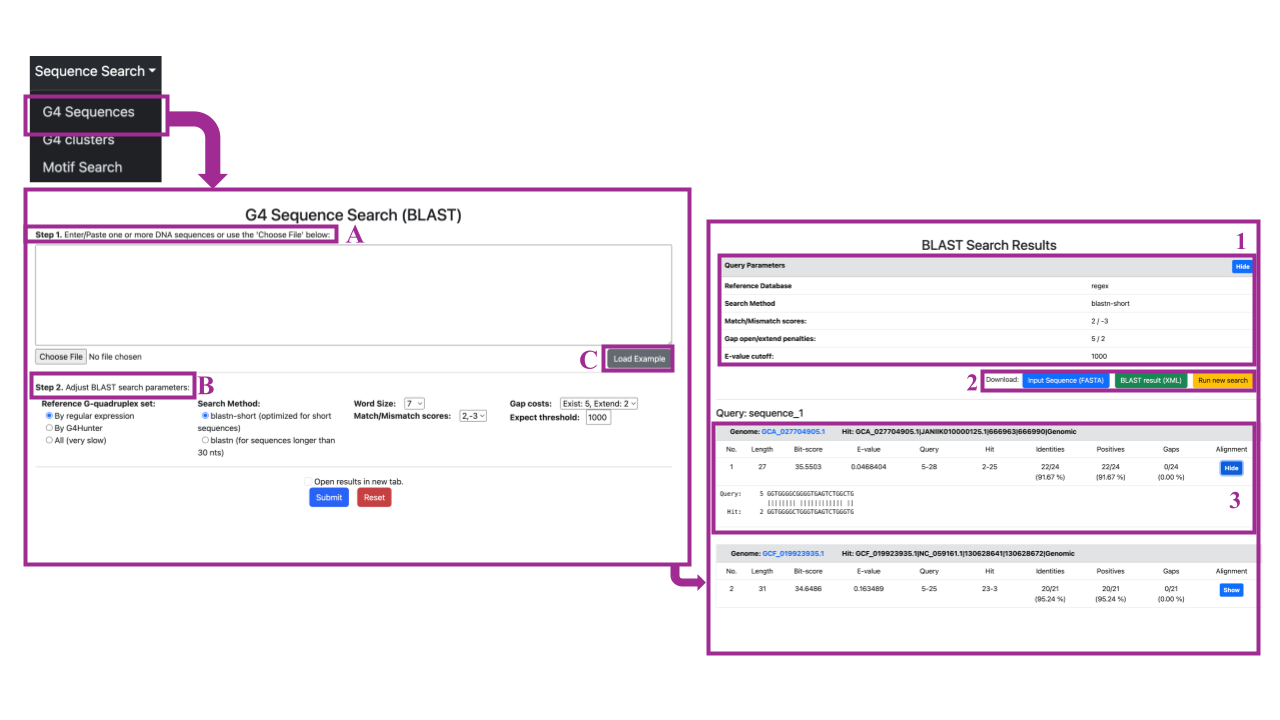
The results will be presented on a page titled "BLAST Search Results":
- Query Parameters Overview:
Here, you can find an overview of the query parameters selected for the search, including the Reference Database, Search Method, Match/Mismatch scores, Gap open/extend penalties, and E-value cutoff.
2.Download and Navigation Options:
You can download the Input Sequence you provided for the search in FASTA format by clicking the blue button labeled "Input Sequence (FASTA)". You can download the BLAST results in XML format by clicking the green button labeled "BLAST (XML)". To return to the search panel, click the yellow button labeled "Run new search"
- Detailed results:
Scrolling down, you will find a table for each hit of each query sequence from the input, displaying all BLAST results based on the selected parameters. The upper gray line shows the genome accession assembly in which the query is aligned and the specific G quadruplex hit in this genome (genome assembly accession | chromosome | start position | end position | type – Genomic/CDS/RNA).
The second line contains alignment details such as the number of the alignment (No), length of the alignment, Bit score, E-value, Query start and end positions, Hit start and end positions, identities score, positives, and gaps.
By clicking the blue button labeled "Show" under the alignment, a new table will be presented with the detailed alignment between the query and the hit. You can hide the alignment by clicking on “Hide” button.
G4 Cluster Search (HMMER)
Selecting Sequence Search → G4 Clusters will redirect you to the "G4 Cluster Search (HMM)" page. Here, you can search by structure in two steps:
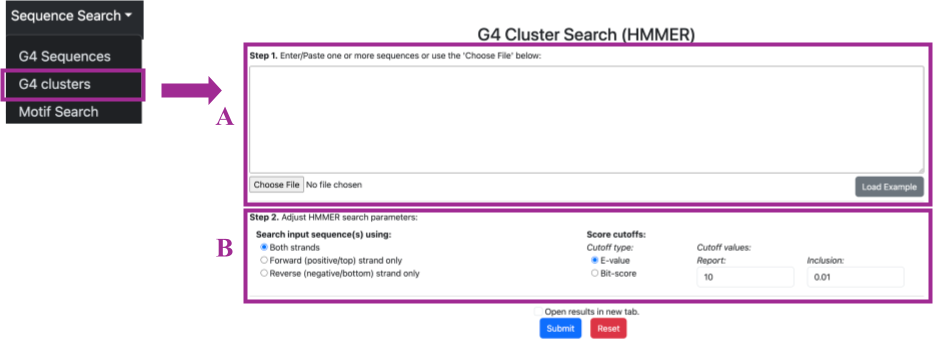
- Step 1 (A): You can either input or paste one or more DNA sequences, or upload a file by clicking the "Choose File" button at the bottom of the search box.
- Step 2 (B): You can adjust HMMER search parameters by selecting the following options:
- Strand selection: In an HMMER search, the search input sequence can be analyzed against one or both strands of the DNA. This is essential for comprehensive sequence analysis because DNA is double-stranded, and genes or motifs can be present on either strand. Here, you can select whether the HMMER search will use both strands, the forward strand, or the reverse strand.
- Cutoff type: The cutoff types are criteria used to determine which hits are considered significant and should be reported in the results. You can filter your results either by E-value or bit-score.
- Cutoff values: Report value and inclusion value: After selecting the cutoff type, you can define the cutoff values by entering the numbers in the respective boxes. The report value determines which hits are displayed in the output, while the inclusion value determines which hits are considered significant and included in the final results.
You can also try examples by clicking on the grey button labeled Load Example. An example sequence will automatically load into the box, allowing you to experiment with different search parameters and become familiar with the search panel.
After the sequence of interest has been loaded in the search panel and the search parameters have been defined, you can submit your selections by clicking the blue Submit button at the bottom of the page, which will redirect you to the results page. If you selected the Open results in a new tab option, your results will be displayed in a new tab. By clicking the red Reset button, you can clear all your selections and start a new search.
The results appear in the same format as the G4 Sequence (BLAST) search.
Motif Search
In the Sequence Search → Motif Search section of the Sequence Search, you can search for specific motifs in two steps:
-
Step 1: Enter the motif in the search panel using the specified format. For G quadruplexes, this typically involves a guanine-rich region with the potential to form G-tetrads. The loops between the G residues are flexible regions that can vary in length and sequence.
-
Step 2: Define search parameters:
- Motif Type: Choose between regular expression or IUPAC sequences. This will depends on the way you select to represent your motif.
- Regular expression (REGEX) motifs are formed using a specialized set of sequence of characters that specifies a match pattern in text. They are complex to write and comprehend, but can create a specialized motif in terms of sequence content, length, and variability and, therefore, capture sequence hits more accurately.
- IUPAC sequences are sequence strings that are formed using the generalized IUPAC nucleotide alphabet. This includes the standard DNA letters A, T, G, C for adenine, thymine, guanine and cytosine, as well as a number of other letters designating positions with variability, e.g. R for A or G, Y for T or C, etc. You can find the complete IUPAC alphabet here.
- Reference G Quadruplex Set: Select the reference G quadruplex set if you want to search against G4 sequences detected only by G4 Hunter, only by regular expression, or both (although this option may be slower).
- Motif Type: Choose between regular expression or IUPAC sequences. This will depends on the way you select to represent your motif.
An example sequence motif, formatted as a regular expression and as an IUPAC sequence, is shown in the table below:
| Regular Expression | IUPAC Sequence | |
| Motif | G{3,}.{1,7}G{3,}.{1,7}G{3,}.{1,7}G{3,} | GGGNNNNNGGGNNGGGNGGG |
When you are ready, you can submit your selections by clicking the blue Submit button at the bottom of the page, which will redirect you to the results page. If you selected the Open results in a new tab option, your results will be displayed in a new tab. By clicking the red Reset button, you can clear all your selections and start a new search.
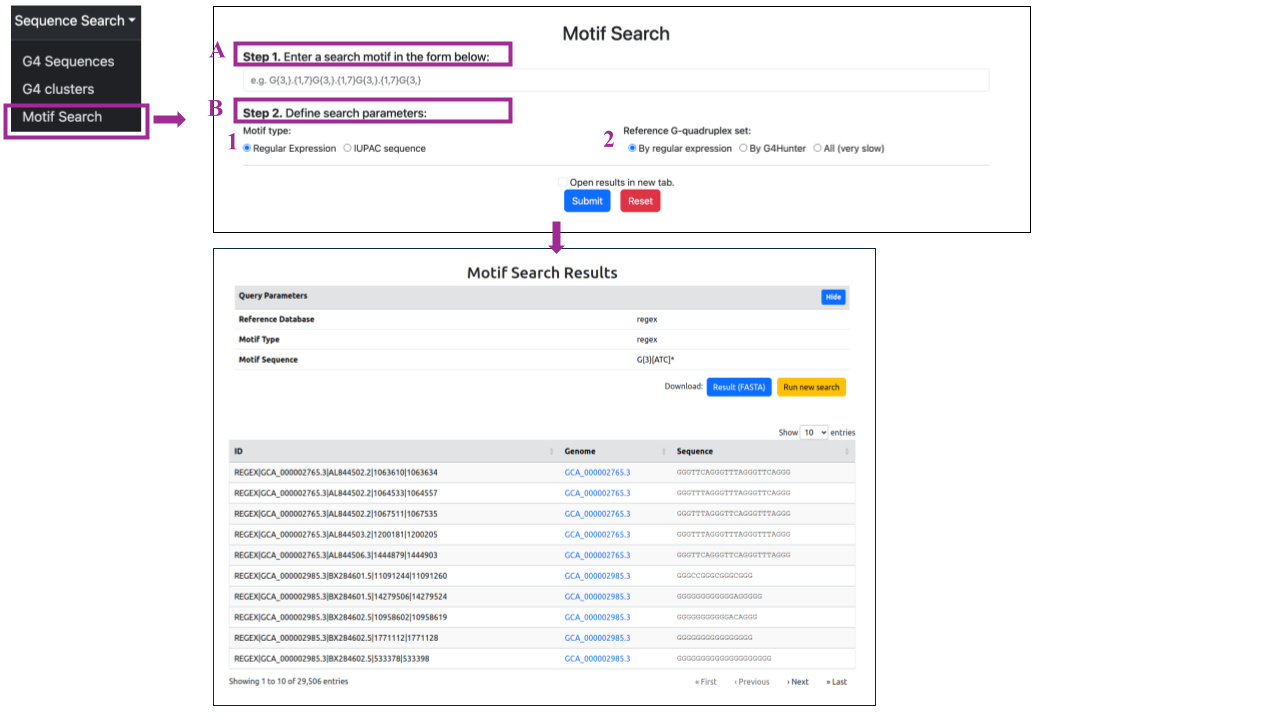
The results will be presented on a page titled "Motif Search Results":
- Query Parameters Overview:
Here, you can find an overview of the query parameters selected for the search, including the Reference Database, Search Method, Match/Mismatch scores, Gap open/extend penalties, and E-value cutoff.
- Download and Navigation Options:
You can download the results in the FASTA format by clicking the blue button labeled "Result (FASTA)". To return to the search panel, click the yellow button labeled "Run new search"
- Detailed results:
Scrolling down, you will find a table for each hit of each query sequence from the input, displaying all results based on the selected parameters. The table contains a column with the sequence header, the corresponding genome and, finally, the G4 sequence containing the queried motif.